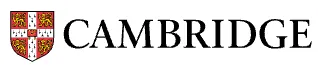About
IT engineer turned data engineer with a side of data scientist. Currently working as a system developer at Lund University, with modern web apps. Enjoys creating order from chaos and finding ways to explain complex concepts in simple terms. A focus on ensuring reproducibility and scalability, while also considering presentation and usability. Outside of data/software work a musician in a number of bands and orchestras, and has too many hobbies: 3D printing, board games, leatherworking, tabletop RPGs, knitting, playing squash, running, and many more. Loves to dig deep in new fields and soak up knowledge.
Posts
Experience
LTH IT, Faculty of Engineering Lund University
System Developer
Jan 2025 - Current

Lund University
Bioinformatician, Cancer and Stem Cell Research
Sep 2019 - Dec 2024

- Developmental Hematopoiesis (David Bryder lab)
- Stem Cells and Leukemia (Göran Karlsson lab)
- Developmental lymphopoiesis and leukemia (Charlotta Böiers lab)
Dealt with quality checking, preprocessing and storing of data, performed analyses, evaluated and chose methods, visualised and communicated results, developed pipelines with reproducibility in mind etc. Worked mostly with sequencing data: RNA, ATAC, CITE (and some CUT&Tag). Performed analyses like (other than 'standard' preprocessing, dimensionality reduction, clustering): finding differentially expressed genes, pseudotime and trajectory inference, mapping and integration, motif analysis. Part of developing two bioinformatic toolkits (Scarf and Nabo). Writing reproducible workflows with Snakemake, Docker, Conda and git/GitHub.
Mikrodust
Software Engineer
Okt 2017 - Dec 2018

Verisure
Research Engineer
Jun 2017 - Aug 2017

Projects
Article: Implications of stress-induced gene expression for hematopoietic stem cell aging studies
Nature Aging.
A study exploring how suboptimal preparation conditions for cells that are to be sequenced (have their RNA or DNA read) can trigger stress responses in the cells and cloud conclusions drawn from the data.
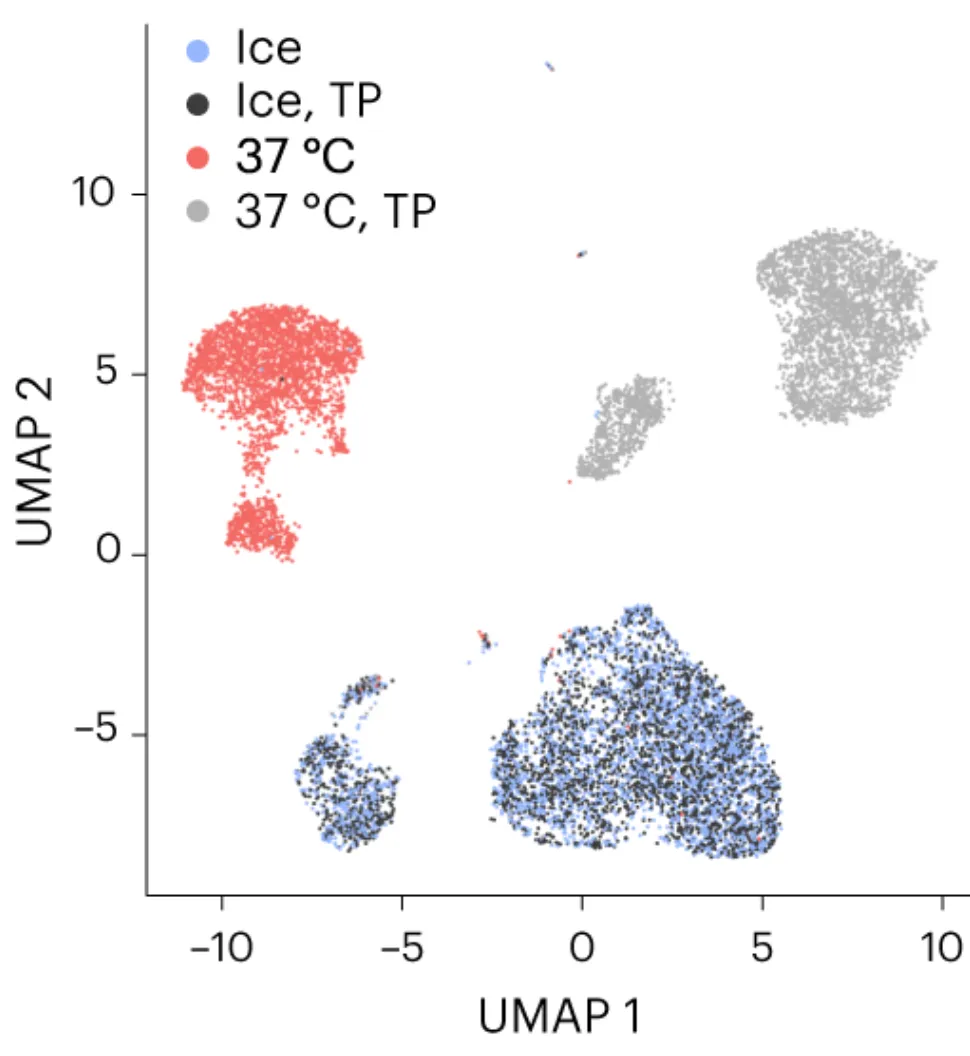
Article: Ex vivo expansion potential of murine hematopoietic stem cells is a rare property only partially predicted by phenotype
eLife
Multiome sequencing (both RNA and DNA from the same cell) of blood stem cells from culture, where we applied a deep learning integration approach and wrote a reproducible pipeline.
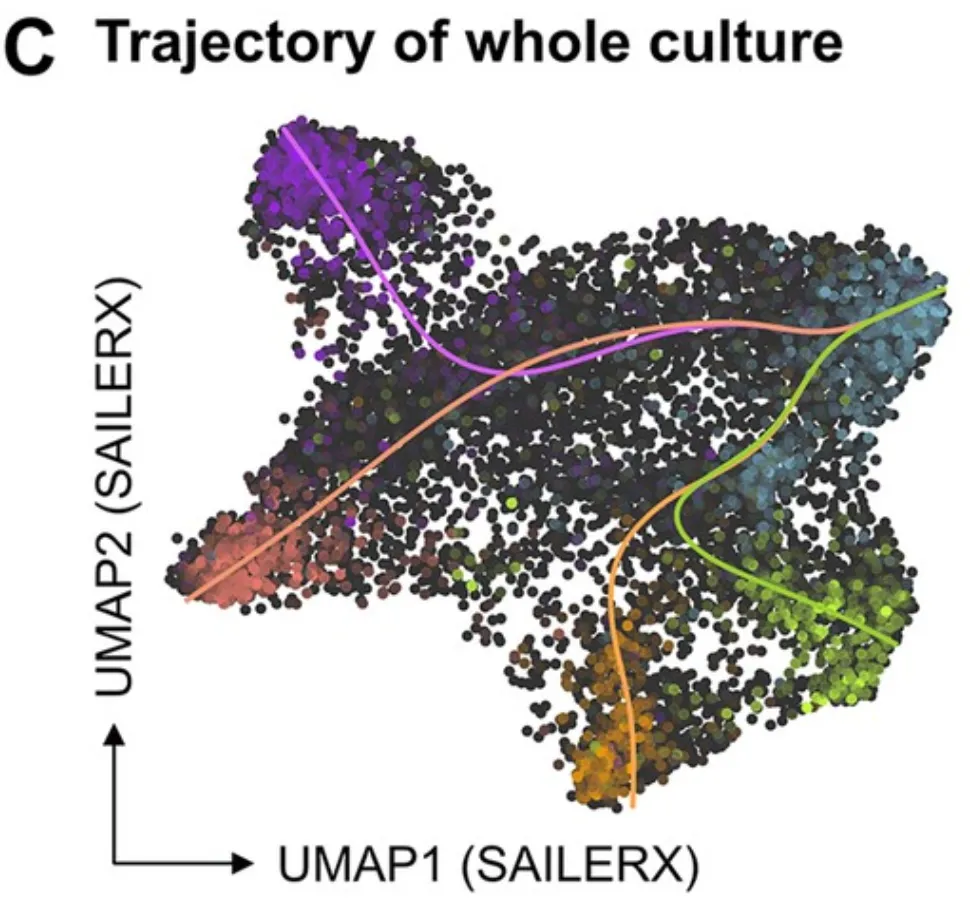
Article: Single-cell multiomics of human fetal hematopoiesis define a developmental-specific population and a fetal signature
Blood Advances
Exploring the differences between fetal and adult blood stem cells, to better combat e.g. childhood leukemia. I did analysis and unified different authors' analyses into a single, reproducible pipeline.

Article: Scarf enables a highly memory-efficient analysis of large-scale single-cell genomics data
Nature Communications
A Python package for performing single-cell sequencing analysis in a memory-efficient way (chunking). I wrote documentation and tests, was part of dogfooding and subsequent bug fixing and improvements, and advised on the design.
Education
Lund University, Faculty of Engineering (LTH)
Master of Science in Engineering; Information and Communication Engineering Technologies
Jan 2016 - Jun 2019 (pt. 2)

- Software engineering, mathematics, interaction design, control theory, physics etc.
- ML/AI (e.g. deep learning, NLP), VR/AR, computer and web security, cognitive robotics etc.
Technische Universität München (TUM)
Exchange studies (ERASMUS)
Oct 2018 - Mar 2019

Lund University, Department of Psychology
Psychology
Sep 2015 - Jan 2016

Lund University, Faculty of Engineering (LTH)
Master of Science in Engineering; Information and Communication Engineering Technologies
Sep 2012 - Jan 2015 (pt. 1)

Musikkonservatoriet Falun
Classical Percussion
Sep 2011 - Jun 2012

Sjöviks Folkhögskola
Classical Percussion
Sep 2010 - Jun 2011

Certifications
Miscellaneous

Discokollektivet
A disco band.

Lunds Brassband
A brass band.